- Research Article
- Open access
- Published:
Analysis of Free Energy Signals Arising from Nucleotide Hybridization Between rRNA and mRNA Sequences during Translation in Eubacteria
EURASIP Journal on Bioinformatics and Systems Biology volume 2006, Article number: 23613 (2006)
Abstract
A decoding algorithm is tested that mechanistically models the progressive alignments that arise as the mRNA moves past the rRNA tail during translation elongation. Each of these alignments provides an opportunity for hybridization between the single-stranded,  -terminal nucleotides of the 16S rRNA and the spatially accessible window of mRNA sequence, from which a free energy value can be calculated. Using this algorithm we show that a periodic, energetic pattern of frequency 1/3 is revealed. This periodic signal exists in the majority of coding regions of eubacterial genes, but not in the non-coding regions encoding the 16S and 23S rRNAs. Signal analysis reveals that the population of coding regions of each bacterial species has a mean phase that is correlated in a statistically significant way with species (
-terminal nucleotides of the 16S rRNA and the spatially accessible window of mRNA sequence, from which a free energy value can be calculated. Using this algorithm we show that a periodic, energetic pattern of frequency 1/3 is revealed. This periodic signal exists in the majority of coding regions of eubacterial genes, but not in the non-coding regions encoding the 16S and 23S rRNAs. Signal analysis reveals that the population of coding regions of each bacterial species has a mean phase that is correlated in a statistically significant way with species ( ) content. These results suggest that the periodic signal could function as a synchronization signal for the maintenance of reading frame and that codon usage provides a mechanism for manipulation of signal phase.
) content. These results suggest that the periodic signal could function as a synchronization signal for the maintenance of reading frame and that codon usage provides a mechanism for manipulation of signal phase.
References
Frick DN, Richardson CC: DNA primases. Annual Review of Biochemistry 2001, 70: 39-80. 10.1146/annurev.biochem.70.1.39
Rymond BC, Rosbash M: Yeast pre-mRNA splicing. In The Molecular and Cellular Biology of the Yeast Saccharomyces: Vol. II. Gene Expression. Volume 2. Edited by: Jones E, Pringle J, Broach J. New York, NY, USA; 1992:143-192.
Doench JG, Petersen CP, Sharp PA: siRNAs can function as miRNAs. Genes and Development 2003,17(4):438-442. 10.1101/gad.1064703
Shine J, Dalgarno L:The
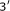 -terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proceedings of the National Academy of Sciences of the United States of America 1974,71(4):1342-1346. 10.1073/pnas.71.4.1342
-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proceedings of the National Academy of Sciences of the United States of America 1974,71(4):1342-1346. 10.1073/pnas.71.4.1342Steitz JA, Jakes K:How ribosomes select initiator regions in mRNA: base pair formation between the
 terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 1975,72(12):4734-4738. 10.1073/pnas.72.12.4734
terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 1975,72(12):4734-4738. 10.1073/pnas.72.12.4734Hui A, de Boer HA: Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 1987,84(14):4762-4766. 10.1073/pnas.84.14.4762
Weiss RB, Dunn DM, Atkins JF, Gesteland RF:Slippery runs, shifty stops, backward steps, and forward hops:
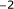 ,
, 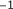 ,
,  ,
,  ,
, 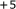 , and
, and  ribosomal frameshifting Cold Spring Harbor Symposia on Quantitative Biology 1987, 52: 687-693.
ribosomal frameshifting Cold Spring Harbor Symposia on Quantitative Biology 1987, 52: 687-693.Weiss RB, Dunn DM, Dahlberg AE, Atkins JF, Gesteland RF:Reading frame switch caused by base-pair formation between the
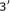 end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO Journal 1988,7(5):1503-1507.
end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO Journal 1988,7(5):1503-1507.Starmer JD: Free2Bind: tools for computing minimum free energy binding between two separate RNA molecules. http://sourceforge.net/projects/free2bind/
Starmer J, Stomp A-M, Vouk MA, Bitzer DL: Predicting Shine-Dalgarno sequence locations exposes genome annotation errors. PLoS Computational Biology 2006,2(5):454-466.
Xia T, SantaLucia J Jr., Burkard ME, et al.: Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 1998,37(42):14719-14735. 10.1021/bi9809425
Jaeger JA, Turner DH, Zuker M: Improved predictions of secondary structures for RNA. Proceedings of the National Academy of Sciences of the United States of America 1989,86(20):7706-7710. 10.1073/pnas.86.20.7706
Mathews DH, Sabina J, Zuker M, Turner DH: Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. Journal of Molecular Biology 1999,288(5):911-940. 10.1006/jmbi.1999.2700
Schluenzen F, Tocilj A, Zarivach R, et al.: Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 2000,102(5):615-623. 10.1016/S0092-8674(00)00084-2
Yusupova GZ, Yusupov MM, Cate JHD, Noller HF: The path of messenger RNA through the ribosome. Cell 2001,106(2):233-241. 10.1016/S0092-8674(01)00435-4
Schurr T, Nadir E, Margalit H: Identification and characterization of E.coli ribosomal binding sites by free energy computation. Nucleic Acids Research 1993,21(17):4019-4023. 10.1093/nar/21.17.4019
Osada Y, Saito R, Tomita M:Analysis of base-pairing potentials between 16S rRNA and
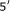 UTR for translation initiation in various prokaryotes. Bioinformatics 1999,15(7-8):578-581.
UTR for translation initiation in various prokaryotes. Bioinformatics 1999,15(7-8):578-581.Thanaraj TA, Pandit MW: An additional ribosome-binding site on mRNA of highly expressed genes and a bifunctional site on the colicin fragment of 16S rRNA from Escherichia coli: important determinants of the efficiency of translation-initiation. Nucleic Acids Research 1989,17(8):2973-2985. 10.1093/nar/17.8.2973
Lithwick G, Margalit H: Hierarchy of sequence-dependent features associated with prokaryotic translation. Genome Research 2003,13(12):2665-2673. 10.1101/gr.1485203
Lee K, Holland-Staley CA, Cunningham PR: Genetic analysis of the Shine-Dalgarno interaction: selection of alternative functional mRNA-rRNA combinations. RNA 1996,2(12):1270-1285.
Komarova AV, Tchufistova LS, Supina EV, Boni IV:Extensive complementarity of the Shine-Dalgarno region and
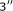 -end of 16S rRNA is inefficient for translation in vivo. Russian Journal of Bioorganic Chemistry 2001,27(4):248-255. 10.1023/A:1011356520576
-end of 16S rRNA is inefficient for translation in vivo. Russian Journal of Bioorganic Chemistry 2001,27(4):248-255. 10.1023/A:1011356520576Ma J, Campbell A, Karlin S: Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. Journal of Bacteriology 2002,184(20):5733-5745. 10.1128/JB.184.20.5733-5745.2002
Oppenheim AV, Schafer RW: Digital Signal Processing. 1st edition. Prentice-Hall, Englewood Cliffs, NJ, USA; 1975.
Brockwell PJ, Davis RA: Time Series: Theory and Methods. 2nd edition. Springer, New York, NY, USA; 1991.
Kay SM: Fundamentals of Statistical Signal Processing, Vol. I: Estimation Theory. Prentice-Hall, Englewood Cliffs, NJ, USA; 1993.
Brocklebank JC, Dickey DA: SAS for Forecasting Time Series. 2nd edition. John Wiley & Sons, New York, NY, USA; 2003.
Lio P, Ruffo S, Buiatti M:Third codon
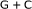 periodicity as a possible signal for an "Internal" selective constraint. Journal of Theoretical Biology 1994,171(2):215-223. 10.1006/jtbi.1994.1225
periodicity as a possible signal for an "Internal" selective constraint. Journal of Theoretical Biology 1994,171(2):215-223. 10.1006/jtbi.1994.1225D'Onofrio G, Bernardi G: A universal compositional correlation among codon positions. Gene 1992,110(1):81-88. 10.1016/0378-1119(92)90447-W
Cocho G, Rius JL: Structural constraints and gene dynamics. Rivista di Biologia - Biology Forum 1989,82(3-4):344-345, 416–417.
Gouy M, Gautier C: Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Research 1982,10(22):7055-7074. 10.1093/nar/10.22.7055
Trifonov EN: Translation framing code and frame-monitoring mechanism as suggested by the analysis of mRNA and 16S rRNA nucleotide sequences. Journal of Molecular Biology 1987,194(4):643-652. 10.1016/0022-2836(87)90241-5
Ponnala L, Barnes T, Bitzer DL, Vouk MA, Stomp A-M: A signal processing-based model for analyzing programmed frameshifts. Proceedings of IEEE International Workshop on Genomic Signal Processing and Statistics (GENSIPS '05), Newport, RI, USA, May 2005
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ponnala, L., Stomp, AM., Bitzer, D.L. et al. Analysis of Free Energy Signals Arising from Nucleotide Hybridization Between rRNA and mRNA Sequences during Translation in Eubacteria. J Bioinform Sys Biology 2006, 23613 (2006). https://doi.org/10.1155/BSB/2006/23613
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1155/BSB/2006/23613
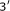 -terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proceedings of the National Academy of Sciences of the United States of America 1974,71(4):1342-1346. 10.1073/pnas.71.4.1342
-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proceedings of the National Academy of Sciences of the United States of America 1974,71(4):1342-1346. 10.1073/pnas.71.4.1342 terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 1975,72(12):4734-4738. 10.1073/pnas.72.12.4734
terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 1975,72(12):4734-4738. 10.1073/pnas.72.12.4734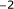 ,
, 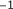 ,
,  ,
,  ,
, 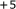 , and
, and  ribosomal frameshifting Cold Spring Harbor Symposia on Quantitative Biology 1987, 52: 687-693.
ribosomal frameshifting Cold Spring Harbor Symposia on Quantitative Biology 1987, 52: 687-693.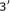 end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO Journal 1988,7(5):1503-1507.
end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO Journal 1988,7(5):1503-1507.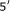 UTR for translation initiation in various prokaryotes. Bioinformatics 1999,15(7-8):578-581.
UTR for translation initiation in various prokaryotes. Bioinformatics 1999,15(7-8):578-581.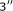 -end of 16S rRNA is inefficient for translation in vivo. Russian Journal of Bioorganic Chemistry 2001,27(4):248-255. 10.1023/A:1011356520576
-end of 16S rRNA is inefficient for translation in vivo. Russian Journal of Bioorganic Chemistry 2001,27(4):248-255. 10.1023/A:1011356520576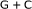 periodicity as a possible signal for an "Internal" selective constraint. Journal of Theoretical Biology 1994,171(2):215-223. 10.1006/jtbi.1994.1225
periodicity as a possible signal for an "Internal" selective constraint. Journal of Theoretical Biology 1994,171(2):215-223. 10.1006/jtbi.1994.1225