- Research Article
- Open access
- Published:
Inferring Time-Varying Network Topologies from Gene Expression Data
EURASIP Journal on Bioinformatics and Systems Biology volume 2007, Article number: 51947 (2007)
Abstract
Most current methods for gene regulatory network identification lead to the inference of steady-state networks, that is, networks prevalent over all times, a hypothesis which has been challenged. There has been a need to infer and represent networks in a dynamic, that is, time-varying fashion, in order to account for different cellular states affecting the interactions amongst genes. In this work, we present an approach, regime-SSM, to understand gene regulatory networks within such a dynamic setting. The approach uses a clustering method based on these underlying dynamics, followed by system identification using a state-space model for each learnt cluster—to infer a network adjacency matrix. We finally indicate our results on the mouse embryonic kidney dataset as well as the T-cell activation-based expression dataset and demonstrate conformity with reported experimental evidence.
References
Rangel C, Angus J, Ghahramani Z, et al.: Modeling T-cell activation using gene expression profiling and state-space models. Bioinformatics 2004, 20(9):1361-1372. 10.1093/bioinformatics/bth093
Perrin B-E, Ralaivola L, Mazurie A, Bottani S, Mallet J, D'Alché-Buc F: Gene networks inference using dynamic Bayesian networks. Bioinformatics 2003, 19(2):II138-II148. 10.1093/bioinformatics/btg1071
Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M: Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 2004, 431(7006):308-312. 10.1038/nature02782
Sontag E, Kiyatkin A, Kholodenko BN: Inferring dynamic architecture of cellular networks using time series of gene expression, protein and metabolite data. Bioinformatics 2004, 20(12):1877-1886. 10.1093/bioinformatics/bth173
Kim S, Li H, Russ D, et al.: Context-sensitive probabilistic Boolean networks to mimic biological regulation. Proceedings of Oncogenomics, Phoenix, Ariz, USA, January-February 2003
Li H, Wood CL, Liu Y, Getchell TV, Getchell ML, Stromberg AJ: Identification of gene expression patterns using planned linear contrasts. BMC Bioinformatics 2006, 7: 245. 10.1186/1471-2105-7-245
Figueiredo MAT, Jain AK: Unsupervised learning of finite mixture models. IEEE Transactions on Pattern Analysis and Machine Intelligence 2002, 24(3):381-396. 10.1109/34.990138
Stuart RO, Bush KT, Nigam SK: Changes in gene expression patterns in the ureteric bud and metanephric mesenchyme in models of kidney development. Kidney International 2003, 64(6):1997-2008. 10.1046/j.1523-1755.2003.00383.x
Khandekar M, Suzuki N, Lewton J, Yamamoto M, Engel JD: Multiple, distant Gata2 enhancers specify temporally and tissue-specific patterning in the developing urogenital system. Molecular and Cellular Biology 2004, 24(23):10263-10276. 10.1128/MCB.24.23.10263-10276.2004
Golyandina N, Nekrutkin V, Zhigljavsky A: Analysis of Time Series Structure—SSA and Related Techniques. Chapman & Hall/CRC, New York, NY, USA; 2001.
Moskvina V, Zhigljavsky A: An algorithm based on singular spectrum analysis for change-point detection. Communications in Statistics Part B: Simulation and Computation 2003, 32(2):319-352. 10.1081/SAC-120017494
Schwab K, Patterson LT, Aronow BJ, Luckas R, Liang H-C, Potter SS: A catalogue of gene expression in the developing kidney. Kidney International 2003, 64(5):1588-1604. 10.1046/j.1523-1755.2003.00276.x
Zhou Y, Lim K-C, Onodera K, et al.: Rescue of the embryonic lethal hematopoietic defect reveals a critical role for GATA-2 in urogenital development. The EMBO Journal 1998, 17(22):6689-6700. 10.1093/emboj/17.22.6689
Challen GA, Martinez G, Davis MJ, et al.: Identifying the molecular phenotype of renal progenitor cells. Journal of the American Society of Nephrology 2004, 15(9):2344-2357. 10.1097/01.ASN.0000136779.17837.8F
NCBI Pubmed[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi]
Zadeh HH, Tanavoli S, Haines DD, Kreutzer DL: Despite large-scale T cell activation, only a minor subset of T cells responding in vitro to Actinobacillus actinomycetemcomitans differentiate into effector T cells. Journal of Periodontal Research 2000, 35(3):127-136. 10.1034/j.1600-0765.2000.035003127.x
Ghahramani Z, Hinton GE: Parameter estimation for linear dynamical systems. University of Toronto, Toronto, Ontario, Canada; 1996.
Shumway RH, Stoffer DS: Time Series Analysis and Applications, Springer Texts in Statistics. Springer, New York, NY, USA; 2000.
Effron B: An Introduction to the Bootstrap. Chapman & Hall/CRC, New York, NY, USA; 1993.
Dougherty ER, Kim S, Chen Y: Coefficient of determination in nonlinear signal processing. Signal Processing 2000, 80(10):2219-2235. 10.1016/S0165-1684(00)00079-7
Kim S, Dougherty ER, Bittner ML, et al.: General nonlinear framework for the analysis of gene interaction via multivariate expression arrays. Journal of Biomedical Optics 2000, 5(4):411-424. 10.1117/1.1289142
Opgen-Rhein R, Strimmer K: Using regularized dynamic correlation to infer gene dependency networks from time-series microarray data. Proceedings of the 4th International Workshop on Computational Systems Biology (WCSB '06), Tampere, Finland, June 2006
Hero AO III, Fleury G, Mears AJ, Swaroop A: Multicriteria gene screening for analysis of differential expression with DNA microarrays. EURASIP Journal on Applied Signal Processing 2004, 2004(1):43-52. special issue on genomic signal processing 10.1155/S1110865704310036
Bar-Joseph Z: Analyzing time series gene expression data. Bioinformatics 2004, 20(16):2493-2503. 10.1093/bioinformatics/bth283
Kundaje A, Antar O, Jebara T, Leslie C: Learning regulatory networks from sparsely sampled time series expression data. Columbia University, New York, NY, USA; 2002.
Balmer JE, Blomhoff R: Gene expression regulation by retinoic acid. Journal of Lipid Research 2002, 43(11):1773-1808. 10.1194/jlr.R100015-JLR200
Esquela AF, Lee SE-J: Regulation of metanephric kidney development by growth/differentiation factor 11. Developmental Biology 2003, 257(2):356-370. 10.1016/S0012-1606(03)00100-3
Maeshima A, Yamashita S, Maeshima K, Kojima I, Nojima Y: Activin a produced by ureteric bud is a differentiation factor for metanephric mesenchyme. Journal of the American Society of Nephrology 2003, 14(6):1523-1534. 10.1097/01.ASN.0000067419.86611.21
Mori M, Ghyselinck NB, Chambon P, Mark M: Systematic immunolocalization of retinoid receptors in developing and adult mouse eyes. Investigative Ophthalmology and Visual Science 2001, 42(6):1312-1318.
Lim K-C, Lakshmanan G, Crawford SE, Gu Y, Grosveld F, Engel JD: Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nature Genetics 2000, 25(2):209-212. 10.1038/76080
Mizutani H, May LT, Sehgal PB, Kupper TS: Synergistic interactions of IL-1 and IL-6 in T cell activation. Mitogen but not antigen receptor-induced proliferation of a cloned T helper cell line is enhanced by exogenous IL-6. Journal of Immunology 1989, 143(3):896-901.
Lin J-X, Leonard WJ:The immediate-early gene product Egr-1 regulates the human interleukin- 2 receptor
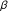 -chain promoter through noncanonical Egr and Sp1 binding sites. Molecular and Cellular Biology 1997, 17(7):3714-3722.
-chain promoter through noncanonical Egr and Sp1 binding sites. Molecular and Cellular Biology 1997, 17(7):3714-3722.Herrgård MJ, Covert MW, Palsson BØ: Reconciling gene expression data with known genome-scale regulatory network structures. Genome Research 2003, 13(11):2423-2434. 10.1101/gr.1330003
Li C, Wong WH: Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proceedings of the National Academy of Sciences of the United States of America 2001, 98(1):31-36. 10.1073/pnas.011404098
Schäfer J, Strimmer K: An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics 2005, 21(6):754-764. 10.1093/bioinformatics/bti062
Rao A, Hero AO III, States DJ, Engel JD: Inference of biologically relevant gene influence networks using the directed information criterion. Proceedings of IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP '06), Toulouse, France, May 2006 2: 1028-1031.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Rao, A., Hero, A.O., States, D.J. et al. Inferring Time-Varying Network Topologies from Gene Expression Data. J Bioinform Sys Biology 2007, 51947 (2007). https://doi.org/10.1155/2007/51947
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1155/2007/51947
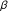 -chain promoter through noncanonical Egr and Sp1 binding sites. Molecular and Cellular Biology 1997, 17(7):3714-3722.
-chain promoter through noncanonical Egr and Sp1 binding sites. Molecular and Cellular Biology 1997, 17(7):3714-3722.