- Research Article
- Open access
- Published:
Splitting the BLOSUM Score into Numbers of Biological Significance
EURASIP Journal on Bioinformatics and Systems Biology volume 2007, Article number: 31450 (2007)
Abstract
Mathematical tools developed in the context of Shannon information theory were used to analyze the meaning of the BLOSUM score, which was split into three components termed as the BLOSUM spectrum (or BLOSpectrum). These relate respectively to the sequence convergence (the stochastic similarity of the two protein sequences), to the background frequency divergence (typicality of the amino acid probability distribution in each sequence), and to the target frequency divergence (compliance of the amino acid variations between the two sequences to the protein model implicit in the BLOCKS database). This treatment sharpens the protein sequence comparison, providing a rationale for the biological significance of the obtained score, and helps to identify weakly related sequences. Moreover, the BLOSpectrum can guide the choice of the most appropriate scoring matrix, tailoring it to the evolutionary divergence associated with the two sequences, or indicate if a compositionally adjusted matrix could perform better.
References
Needleman SB, Wunsch CD: A general method applicable to the search for similarities in the amino acid sequence of two proteins. Journal of Molecular Biology 1970, 48(3):443-453. 10.1016/0022-2836(70)90057-4
McLachlan AD:Tests for comparing related amino-acid sequences. Cytochrome
 and cytochrome
and cytochrome  . Journal of Molecular Biology 1971, 61(2):409-424. 10.1016/0022-2836(71)90390-1
. Journal of Molecular Biology 1971, 61(2):409-424. 10.1016/0022-2836(71)90390-1Sankoff D: Matching sequences under deletion-insertion constraints. Proceedings of the National Academy of Sciences of the United States of America 1972, 69(1):4-6. 10.1073/pnas.69.1.4
Sellers PH: On the theory and computation of evolutionary distances. SIAM Journal on Applied Mathematics 1974, 26(4):787-793. 10.1137/0126070
Waterman MS, Smith TF, Beyer WA: Some biological sequence metrics. Advances in Mathematics 1976, 20(3):367-387. 10.1016/0001-8708(76)90202-4
Dayhoff MO, Schwartz RM, Orcutt BC: A model of evolutionary change in proteins. In Atlas of Protein Sequence and Structure. Volume 5. Edited by: Dayhoff MO. National Biomedical Research Foundation, Washington, DC, USA; 1978:345-352.
Altschul SF: Amino acid substitution matrices from an information theoretic perspective. Journal of Molecular Biology 1991, 219(3):555-565. 10.1016/0022-2836(91)90193-A
Karlin S, Altschul SF: Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proceedings of the National Academy of Sciences of the United States of America 1990, 87(6):2264-2268. 10.1073/pnas.87.6.2264
Henikoff S, Henikoff JG: Amino acid substitution matrices from protein blocks. Proceedings of the National Academy of Sciences of the United States of America 1992, 89(22):10915-10919. 10.1073/pnas.89.22.10915
Feller W: An Introduction to Probability and Its Applications. John Wiley & Sons, New York, NY, USA; 1968.
Yu Y-K, Wootton JC, Altschul SF: The compositional adjustment of amino acid substitution matrices. Proceedings of the National Academy of Sciences of the United States of America 2003, 100(26):15688-15693. 10.1073/pnas.2533904100
Altschul SF: A protein alignment scoring system sensitive at all evolutionary distances. Journal of Molecular Evolution 1993, 36(3):290-300. 10.1007/BF00160485
States DJ, Gish W, Altschul SF: Improved sensitivity of nucleic acid database searches using application-specific scoring matrices. Methods 1991, 3(1):66-70. 10.1016/S1046-2023(05)80165-3
Sunyaev SR, Bogopolsky GA, Oleynikova NV, Vlasov PK, Finkelstein AV, Roytberg MA: From analysis of protein structural alignments toward a novel approach to align protein sequences. Proteins: Structure, Function, and Bioinformatics 2004, 54(3):569-582.
Zachariah MA, Crooks GE, Holbrook SR, Brenner SE: A generalized affine gap model significantly improves protein sequence alignment accuracy. Proteins: Structure, Function, and Bioinformatics 2005, 58(2):329-338.
Shannon CE: A mathematical theory of communication—part I. Bell System Technical Journal 1948, 27: 379-423.
Shannon CE: A mathematical theory of communication—part II. Bell System Technical Journal 1948, 27: 623-656.
Csiszár I, Körner J: Information Theory: Coding Theorems for Discrete Memoryless Systems. Academic Press, New York, NY, USA; 1981.
Schäffer AA, Aravind L, Madden TL, et al.: Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Research 2001, 29(14):2994-3005. 10.1093/nar/29.14.2994
Frommlet F, Futschik A, Bogdan M: On the significance of sequence alignments when using multiple scoring matrices. Bioinformatics 2004, 20(6):881-887. 10.1093/bioinformatics/btg498
Altschul SF, Wootton JC, Gertz EM, et al.: Protein database searches using compositionally adjusted substitution matrices. FEBS Journal 2005, 272(20):5101-5109. 10.1111/j.1742-4658.2005.04945.x
Schäffer AA, Wolf YI, Ponting CP, Koonin EV, Aravind L, Altschul SF: IMPALA: matching a protein sequence against a collection of PSI-BLAST-constructed position-specific score matrices. Bioinformatics 1999, 15(12):1000-1011. 10.1093/bioinformatics/15.12.1000
Rypniewski WR, Perrakis A, Vorgias CE, Wilson KS: Evolutionary divergence and conservation of trypsin. Protein Engineering 1994, 7(1):57-64. 10.1093/protein/7.1.57
Hughes AL: Evolutionary diversification of the mammalian defensins. Cellular and Molecular Life Sciences 1999, 56(1-2):94-103. 10.1007/s000180050010
Bauer F, Schweimer K, Klüver E, et al.:Structure determination of human and murine
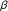 -defensins reveals structural conservation in the absence of significant sequence similarity. Protein Science 2001, 10(12):2470-2479. 10.1110/ps.ps.24401
-defensins reveals structural conservation in the absence of significant sequence similarity. Protein Science 2001, 10(12):2470-2479. 10.1110/ps.ps.24401Tossi A, Sandri L: Molecular diversity in gene-encoded, cationic antimicrobial polypeptides. Current Pharmaceutical Design 2002, 8(9):743-761. 10.2174/1381612023395475
Gennaro R, Zanetti M, Benincasa M, Podda E, Miani M: Pro-rich antimicrobial peptides from animals: structure, biological functions and mechanism of action. Current Pharmaceutical Design 2002, 8(9):763-778. 10.2174/1381612023395394
Selsted ME, Novotny MJ, Morris WL, Tang Y-Q, Smith W, Cullor JS: Indolicidin, a novel bactericidal tridecapeptide amide from neutrophils. Journal of Biological Chemistry 1992, 267(7):4292-4295.
Kullback S: Information Theory and Statistics. Dover, Mineola, NY, USA; 1997.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Fabris, F., Sgarro, A. & Tossi, A. Splitting the BLOSUM Score into Numbers of Biological Significance. J Bioinform Sys Biology 2007, 31450 (2007). https://doi.org/10.1155/2007/31450
Received:
Accepted:
Published:
DOI: https://doi.org/10.1155/2007/31450
 and cytochrome
and cytochrome  . Journal of Molecular Biology 1971, 61(2):409-424. 10.1016/0022-2836(71)90390-1
. Journal of Molecular Biology 1971, 61(2):409-424. 10.1016/0022-2836(71)90390-1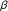 -defensins reveals structural conservation in the absence of significant sequence similarity. Protein Science 2001, 10(12):2470-2479. 10.1110/ps.ps.24401
-defensins reveals structural conservation in the absence of significant sequence similarity. Protein Science 2001, 10(12):2470-2479. 10.1110/ps.ps.24401