Table 1 P53 pathways, with GS size ( ), unadjusted and FDR adjusted
), unadjusted and FDR adjusted  -values (
-values (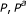 )
)
From: A Hypothesis Test for Equality of Bayesian Network Models
Pathway |
|
|
| Sub | Efr | Liu |
|---|---|---|---|---|---|---|
SA_G1_AND_S_PHASES | 14 |
| .08 | n | y | n |
atmPathway | 19 |
| .08 | n | n | y |
g2Pathway | 23 |
| .08 | n | n | n |
p53Pathway | 16 |
| .08 | y | y | y |
cell_cycle_checkpointII | 10 |
| .08 | n | n | n |
SA_FAS_SIGNALLING | 9 | .002 | .14 | n | n* | n* |
cellcyclePathway | 23 | .002 | .16 | n | n* | n* |
DNA_ | 90 | .003 | .17 | n | n* | n* |
SA_TRKA_RECEPTOR | 16 | .003 | .17 | n | n* | y* |
radiation_sensitivity | 26 | .003 | .17 | y | y* | y* |
ngfPathway | 19 | .004 | .17 | n | y* | n* |
GO_ROS | 23 | .004 | .17 | n | n* | n* |
etsPathway | 16 | .004 | .17 | n | n* | n* |
ck1Pathway | 15 | .006 | .21 | n | n* | n* |
erkPathway | 29 | .007 | .23 | n | n* | n* |
| 18 | .007 | .23 | n | n* | n* |
arfPathway | 13 | .007 | .23 | n | n* | n* |
 ), unadjusted and FDR adjusted
), unadjusted and FDR adjusted  -values (
-values (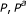 )
)


 .001
.001 .001
.001 .001
.001 .001
.001 .001
.001

 The complete name of DNA_DAMAGE is DNA_DAMAGE_SIGNALLING.
The complete name of DNA_DAMAGE is DNA_DAMAGE_SIGNALLING.  The complete name of MAP00562 is MAP00562_Inositol_phosphate_metabolism. *Inclusion criterion based on control rate of original analysis.
The complete name of MAP00562 is MAP00562_Inositol_phosphate_metabolism. *Inclusion criterion based on control rate of original analysis.